
|
|
The FDA and CDISC have been publishing rules for validating submission datasets (SDTM, SEND, ADaM).
Unfortunately, these rules have been published as pure text (usually in Excel Worksheets), meaning that:
A better way is to publish such rules in an open, vendor-neutral, human-readable but machine-executable format. Such a format is XQuery.
XQuery is an open W3C standard for quering XML documents and databases.
To start with, we implemented almost all of the FDA-SDTM rules, the PMDA-SDTM rules, and the CDISC-SDTM rules (SDTM-IG 3.2) in XQuery, and have started to develop the XQueries for the "CDISC ADaM v1.3 Validation Checks" and for the SDTM rules defined by the "SDTM Validation Subteam".
As XQuery is a query language for XML, you can use and test the rules on Dataset-XML submission files. If you are still working with the outdated SAS Transport 5 format, you can easily transform your SAS Transport 5 files to Dataset-XML files with one of the tools provided by CDISC.
We also make a RESTful web service available for these rules written in XQuery, meaning that:
A presentation at the German-Speaking User Group meeting about the power of XQuery for really open SDTM/ADaM/SEND validation rules (mostly non-technical)
A movie (for techies - no sound yet) about how easy it is to implement ADaM Validation Rules in XQuery
The best is to have your Dataset-XML files, including your define.xml file (v.2.0) in a native XML database such as eXist or BaseX.
The reason is that you can then index your database so that your queries will run considerably faster.
If you don't have a native XML database or do not know how to work with such, you can also just use a file system (i.e. directory). We will explain in a moment how to proceed in that case.
We have implemented the following RESTfull web services (many more will follow)
We will take the rule FDAC050 (Exposure end date may not be after the latest Disposition date) as an example. The screenshots you see come from eXide, the graphical user interface of the eXist native XML database, but you can of course also execute the XQuery rules using any modern application designed for it such as written in Java, C#, C++, ..., or you can easily write your own custom application, using one of the published XQuery libraries such as XQJ (XQuery Processor for Java).
Let us first inspect the first few lines of our XQuery:
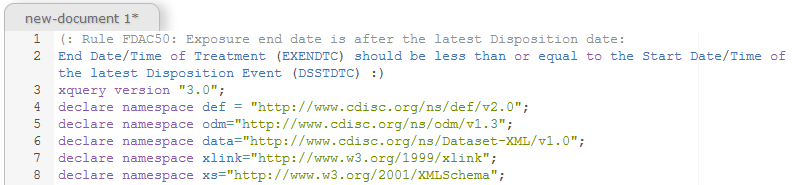
Comments in XQuery start with an "(:" and end with a ":)". So the two first lines are just comments - they are not processed.
Line 3 states with which version of XQuery we are working with - all the rules were written using v.3.0
Lines 4-8 declare all the XML namespaces that we will be working with and their prefixes: you will immediately identify the ODM and the Define-XML namespace, as well as the Dataset-XML namespace.
The following lines are:
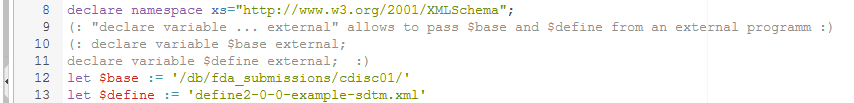
Line 12 defines where the base (directory or database collection) of all our XML documents is. In our case, we stored the XML documents in the eXist native XML database in the collection "/db/fda_submissions/cdisc01". Also the FDA could easily set up a native XML database and store each submission in a different "collection" - that would be much less expensive than the Janus warehouse! In case you work with a normal file system, the statement will be something like: "file:///d:/fda_submissions/cdisc01". It can however also be a URL like: http://mycompany.com/fda_submissions/cdisc01.
Line 13 is very important, as it defines the name of the define.xml file which is assumed to be in the base directory or collection together with all submission files. This is important, as the define.xml is "leading",
i.e. all the information about where the submission files are located, how they are named, what variables with which datatype and length are present, which codelists are present, etc..
This is exactly how it should be the case, a fact otherwise often ignored by other validators
If you have written your own application to execute the XQuery rules, you will probably want to pass the base location and the name of the define.xml as parameters: this is explained (but commented out here) in lines 9-11.
We will later explain how one can set these values from a Java programm.
The following lines are:

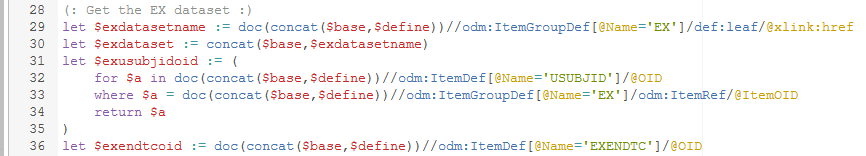
In lines 15, it is read from the define.xml document where the document with data for the DS domain can be found (define.xml is leading!), and as this is a relative path, the full path is generated by concatenating the base and the DS document name.
The following lines are:

This a bit more difficult, as a subquery is used here ...
Essentially what is done here is that the ODM-OIDs of the variables 'USUBJID' and 'DSSTDTC' in the DS dataset are identified as, as you know, datapoints in Dataset-XML are identified with their OIDs,
which are defined in the define.xml file (define.xml is leading!).
So the outcome for $dsusubjoid and $dsstdtcoid will be something like "DS.USUBJID" and "DS.DSSTDTC" depending on how they are defined in the define.xml document.
These two variables will enable to find the data points in the Dataset-XML document.
The following lines are:
The following lines are:
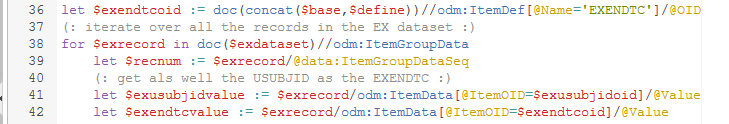
The "for" statement (line 38) iterates over all the records in the EX dataset, where an "ItemGroupData" corresponds to a single record. Within each record, the record number is identified by the Dataset-XML "ItemGroupDataSeq" attribute (line 39).
Lines 41 and 42 then retrieve the value for USUBJID and EXENDTC from the "Value" attribute of the appropriate "ItemData" element (each "ItemData" represents a single data point).
This is followed by:

The following lines:
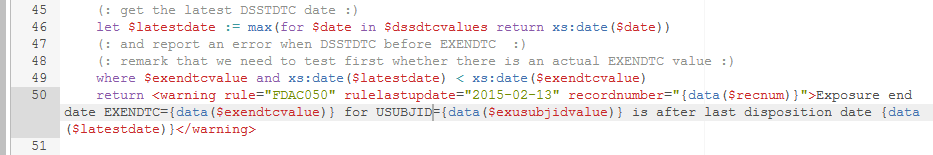
The EXENDTC is compared (as a date) with the value of $latestdate (which is the latest disposition date for the specific subject) in the "where" statement , and if EXENDTC is beyond the latest disposition date, an XML "warning" element is returned, containing information about the record number (in the EX dataset), the observed value for EXENDTC, the USUBJID and the latest disposition date. Remark that also the "last update date" of the rule itself is returned, allowing implementors to see whether the rule (or at least its implementation) were changed.
In our native XML database, the rule executes in about 0.2 seconds for a small submission, in 40 seconds for a large submission.
For more examples with explanations, see our "CDISC Validation Rules in XQuery"
The advantage of having the validation rules as XQuery and using the web service are obvious:
There are many advantages of this approach (vendor neutral, open standard, human- as well as machine-readable, ...) but there are also a few disadvantages:
You will usually want to run the XQueries using a validation softare (similar to OpenCDISC). Writing such validation software is pretty straightforward, and is explained on our "XQuery Implementation" page.
With the first set of rules (CDISC-FDA-PMDA-SDTM/ADaM) implemented as fully transparent, machine-readable and -executable XQuery rules, we have started implementing them in the XML4Pharma SDTM-ETL(TM) mapping software. Also, the rules have been implemented in the open-source "Smart Dataset-XML Viewer". This will enable pharma companies, CROs and service providers to get rid of the currently used validators that have non-transparent rules implemented, with many false-positives, and to which no company-specific rules can be added, at least not in the free version.
Courtesy of XML4Pharma - last update: March 2017