

|
With the "Open Rules for CDISC Standards" everyone, every company, can develop its own software for executing FDA, PMDA and CDISC validation rules,
this as the rules in written in the W3C standardized vendor-neutral XQuery language.
This makes you independent of a single provider (one is well known for the many bugs and "false positives" in its software).
With "Open Rules for CDISC Standards", you can inspect each of the rule implementations, improve them, correct them immediately if necessary (please let us know,
so that we can make an update available to everyone within a few hours - no more waiting until the next release which is next year).
You can even use these rule implementations as a template for your own additional, company-specific rules, and deploy them immediately.
In the next sections, we explain you how you can easily use these rules in your own software, be it Java, C# or any other modern computer language.
The rules require you to have your SDTM, SEND or ADaM datasets in the modern CDISC Dataset-XML format.
The completely outdated SAS Transport 5 (.xpt format) is not supported (this format is not vendor-neutral).
In most cases, you will need to pass two parameters to the software that is executing the rules, which is explained in detail below.
The first parameter is the "location" of where the whole submission (files) is located. This can be:
In some cases there is a third parameter, allowing you to pass a single file or resource. For example, for rules that take somewhat more time to complete, you might want to validate only one file (e.g. "VS.xml") instead of all findings files (all files is the default).
When having downloaded validation rules in XQuery (either using the web service or from the regular download), and inspecting what has been returned, you will notice that the hardcoded location of the define.xml has been commented out (everything between "(:" and ":)" is comment). Instead you will find:

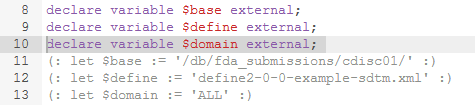
For rules that need to read MedDRA files, you will need to pass the location of your MedDRA folder. The parameter for this is $meddrabase.
For example: $meddrabase='C:\meddra_19_0_english\MedAscii'.
Unfortunately, MedDRA is still propriety and requires a license, and is deployed using 30 year old technology (ASCII files).
You will usually want to run the XQueries using a validation software (similar to OpenCDISC - but better). Writing such validation software is pretty straightforward, but we still like to provide you a "jumpstart".
Here is a Java programm that we developed for testing and which you can use as a base. There are two methods, one for quering Dataset-XML submission files that are stored as files, and the second for quering submission Dataset-XML documents that are in a native XML database (eXist-DB in this case).
Let's go through the steps. First of all the libraries that are needed:
We used SaxonHE9 for parsing and as the XQuery engine, so you will need the saxonhe9.jar and the saxon9-xqj.jar which you can get from the Saxon website
Further you will need the xmldb.jar library, which you can obtain from several sources. If you do already have the eXist native XML database installed, you can find it in directory /lib/core.
if you prefer to use BaseX as native XML database, you can probably also find it in its distribution (I haven't tried yet).
The method 'runXQueryOnFile' is for the case that the Dataset-XML submission files are in a file system:
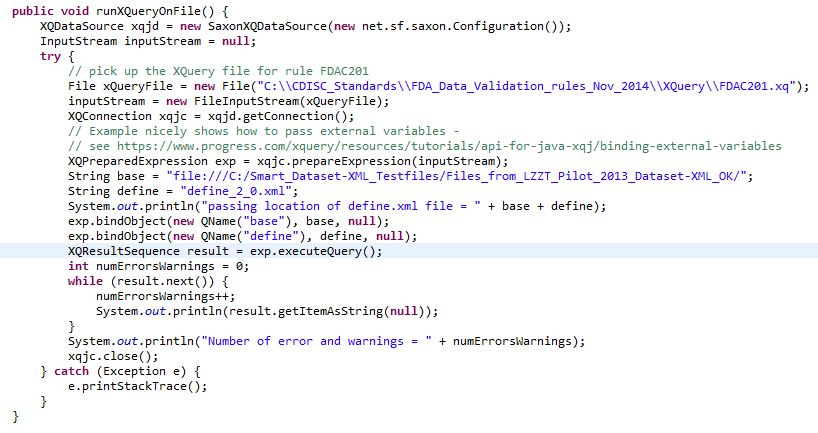
First we tell the system that we want to use the Saxon XQuery engine and an InputStream is defined for the file containing the XQuery - in this case it contains the validation rule FDAC201.
Then the XQuery engine is set up and the contents of the file are put into a "XQPreparedExpression" (which is like a prepared statement in Java-SQL).
The contents of the next lines ("exp.bindObject") pass the location of the define.xml file to the XQuery - also see the $base and $define variables in the XQuery. Remark that the XQuery does not need to know where the other
Dataset-XML files are located, as the location is read from the define.xml document (using "def:leaf xlink:href").
Then the XQuery is executed ("exp.executeQuery();") and an iteration over the set with results is performed. An example output is e.g.:

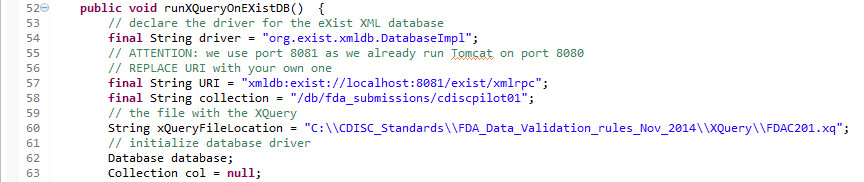
The second method "runXQueryOnEXistDB" demonstrates how to proceed when the define.xml and Dataset-XML files are stored in a native XML database (in this case eXist-DB).
The first lines of the method are:
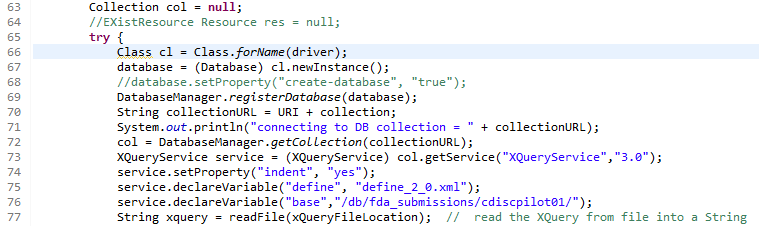
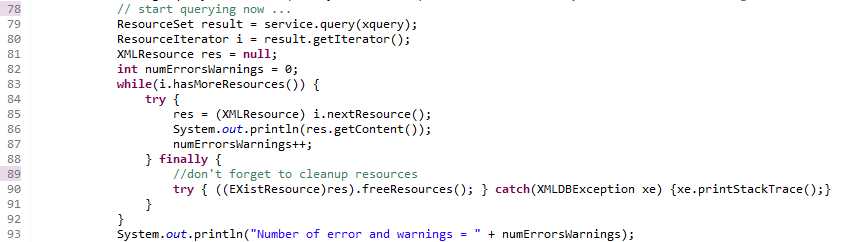
A typical example output is:

Remark that this code is a "quick and dirty" code, not optimized for performance at all. But I think it is a good start anyway.
If you need some help with one these, please do not hesitate to contact us (info-at-xml4pharma.com).
Courtesy of XML4Pharma - last update: March 2017